QFRAGS: An Accurate High-Performance Automatic Molecular Fragmentation for Quantum Chemistry (Best Poster Finalist)
Monday, May 13, 2024 3:00 PM to Wednesday, May 15, 2024 4:00 PM · 2 days 1 hr. (Europe/Berlin)
Foyer D-G - 2nd floor
Research Poster
Chemistry and Materials ScienceNovel Algorithms
Information
Poster is on display and will be presented at the poster pitch session.
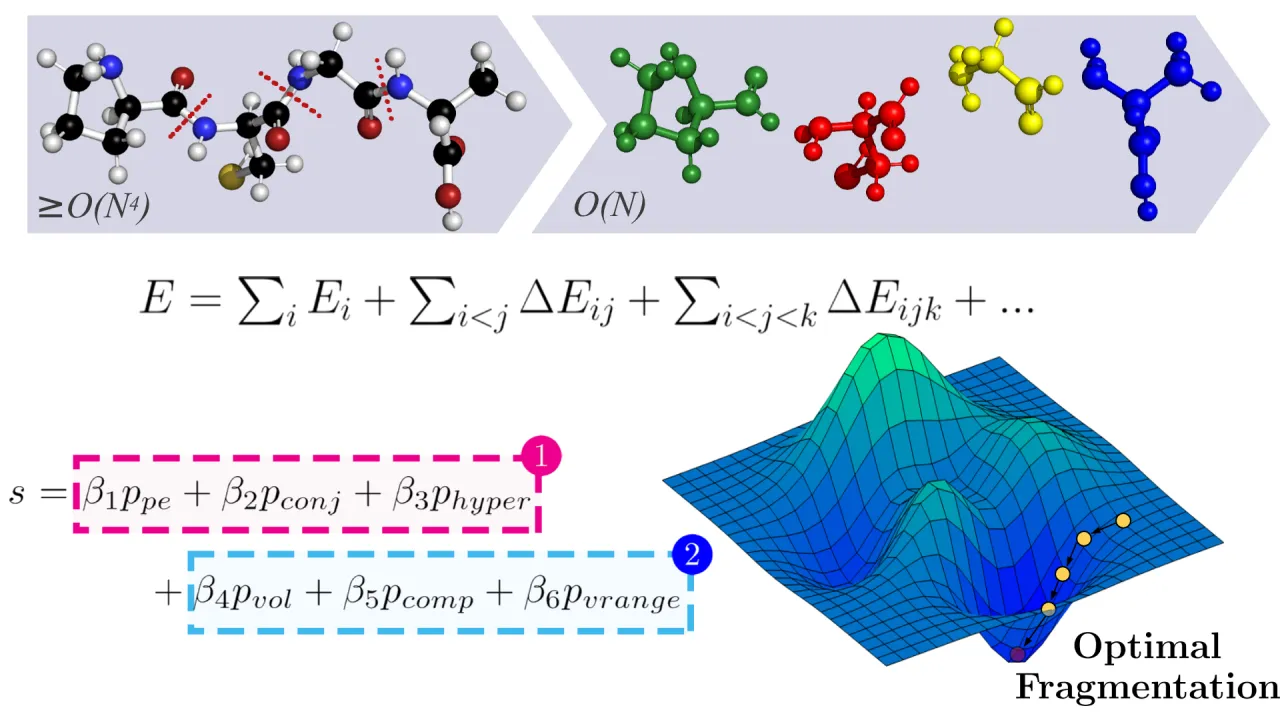
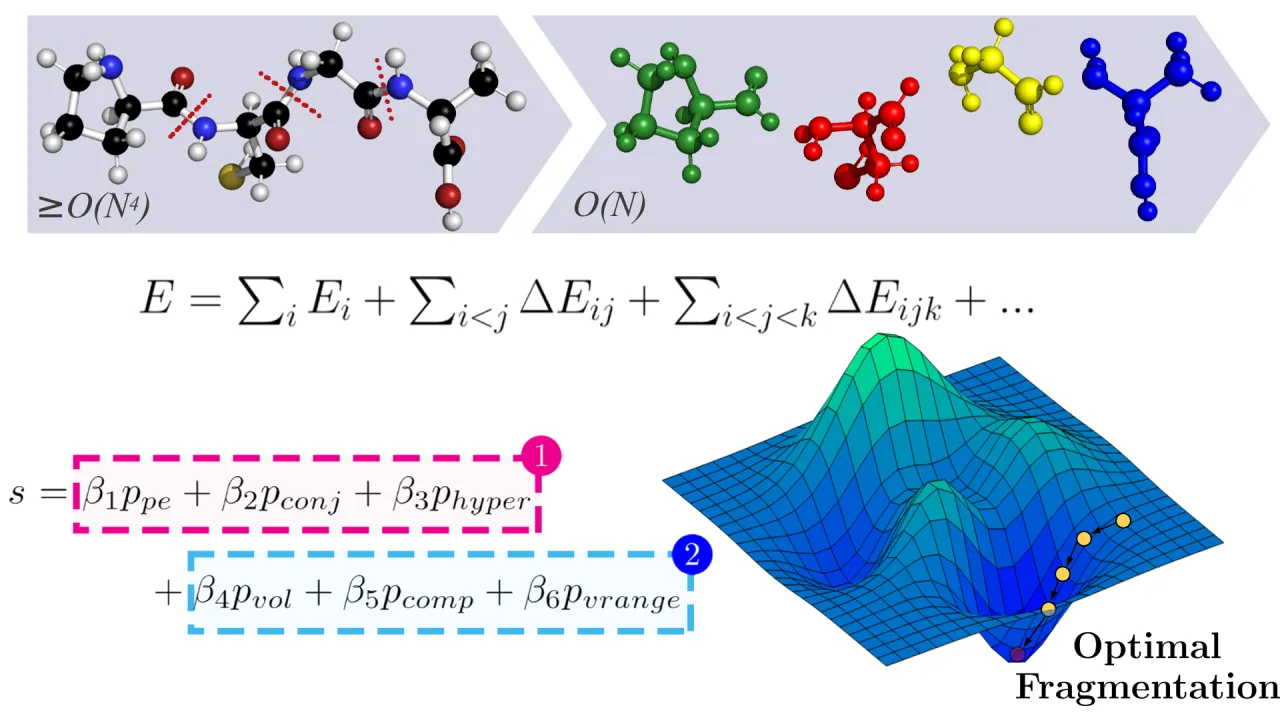
The chemico-physical characterisation of large, dynamic molecular systems is a key challenge in computational chemistry. Modern quantum chemical (QC) calculations provide highly accurate computational models of the chemico-physical behaviour of matter and therefore offer a potential resolution for the characterisation of large molecular systems. However, the computational time required by accurate QC algorithms grows rapidly (>=O(N^4)) with system size, limiting their applicability to systems containing less than a few hundred atoms. Furthermore, underpinning QC algorithms are typically incapable of exploiting the massive parallelism of supercomputer architectures, exacerbating the problem. Molecular fragmentation has emerged as a successful method to reduce the time complexity of underpinning QC algorithms to O(N) and enhance algorithmic parallelisability. However, such fragmentation approaches are not widely adopted due to a dearth of automated fragmentation methods; most involve manual fragmentation and furthermore, none are optimised for supercomputer architecture, prolonging time-to-solution. We present QFRAGS, an accurate high-performance automated fragmentation algorithm capable of handling molecular systems of thousands of atoms. QFRAGS employs a scoring function to define the quality of fragmentation and a corresponding optimisation procedure to attain high quality fragments. A hybrid message passing and shared memory parallel programming (MPI-OpenMP) model was adopted to exploit CPUs across multiple nodes to accelerate the optimisation of the scoring function. Significant speedup was achieved by implementing schemes to efficiently expose parallelism, and employing MPI and OpenMP for parallelism on CPUs. Finally, we show the capability of the fragmentation procedure at generating fragments of a user-specified size and its accuracy.
Contributors:
The chemico-physical characterisation of large, dynamic molecular systems is a key challenge in computational chemistry. Modern quantum chemical (QC) calculations provide highly accurate computational models of the chemico-physical behaviour of matter and therefore offer a potential resolution for the characterisation of large molecular systems. However, the computational time required by accurate QC algorithms grows rapidly (>=O(N^4)) with system size, limiting their applicability to systems containing less than a few hundred atoms. Furthermore, underpinning QC algorithms are typically incapable of exploiting the massive parallelism of supercomputer architectures, exacerbating the problem. Molecular fragmentation has emerged as a successful method to reduce the time complexity of underpinning QC algorithms to O(N) and enhance algorithmic parallelisability. However, such fragmentation approaches are not widely adopted due to a dearth of automated fragmentation methods; most involve manual fragmentation and furthermore, none are optimised for supercomputer architecture, prolonging time-to-solution. We present QFRAGS, an accurate high-performance automated fragmentation algorithm capable of handling molecular systems of thousands of atoms. QFRAGS employs a scoring function to define the quality of fragmentation and a corresponding optimisation procedure to attain high quality fragments. A hybrid message passing and shared memory parallel programming (MPI-OpenMP) model was adopted to exploit CPUs across multiple nodes to accelerate the optimisation of the scoring function. Significant speedup was achieved by implementing schemes to efficiently expose parallelism, and employing MPI and OpenMP for parallelism on CPUs. Finally, we show the capability of the fragmentation procedure at generating fragments of a user-specified size and its accuracy.
Contributors:
Format
On-site

