Developing a Parallel Algorithm to Simulate the Merging of Proteins Using a Mesh-based Approach
Monday, May 13, 2024 3:00 PM to Wednesday, May 15, 2024 4:00 PM · 2 days 1 hr. (Europe/Berlin)
Foyer D-G - 2nd floor
Women in HPC
Bioinformatics and Life SciencesEngineeringNovel Algorithms
Information
Poster is on display and will be presented at the poster pitch session.
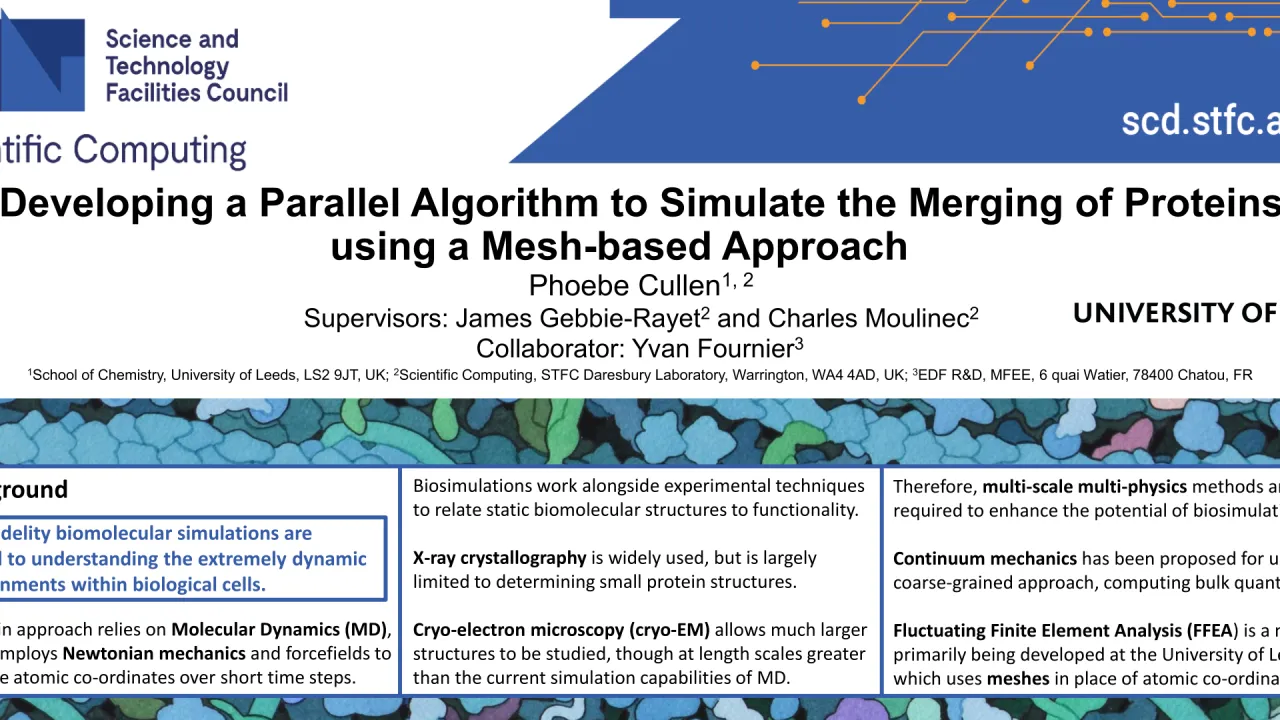
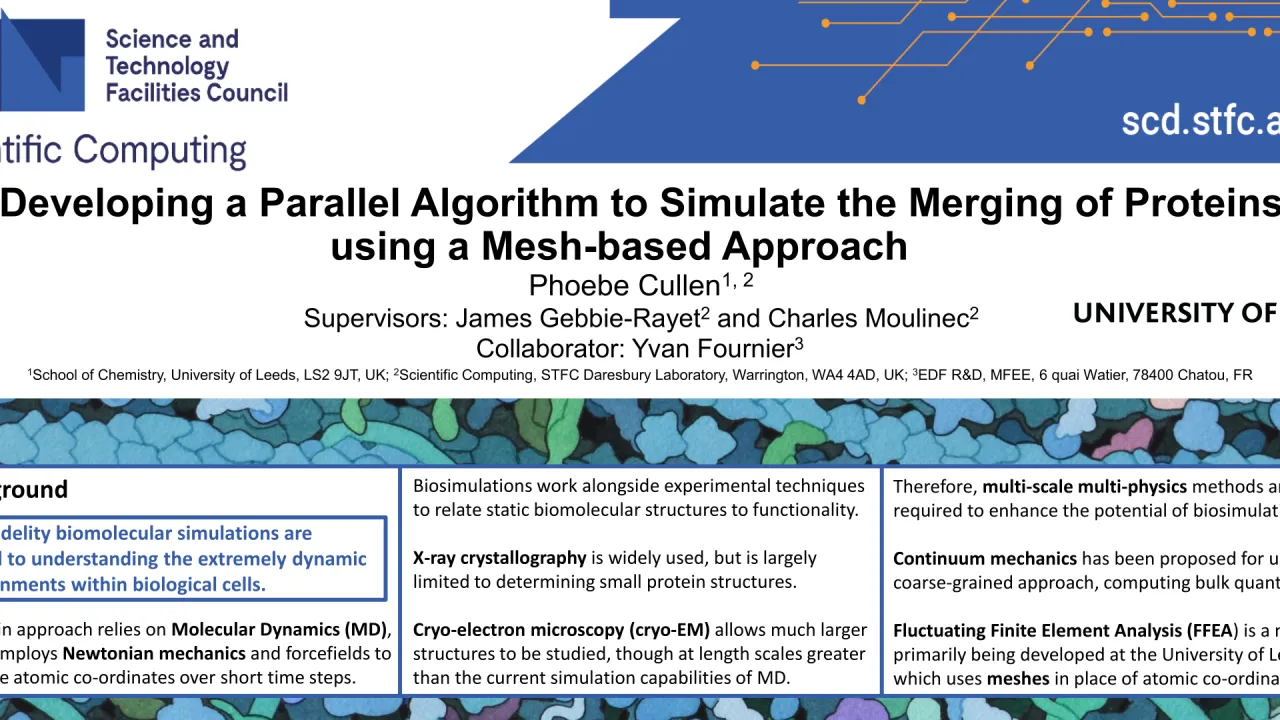
Biomolecular simulations are crucial to understanding the complex and highly dynamic environments within biological cells. A popular approach relies on Molecular Dynamics (MD), which employs Newtonian mechanics and forcefields to compute atomic co-ordinates over short time steps. Biosimulations work in conjunction with experimental techniques, providing dynamic information to relate static biomolecular structures to functionality [1]. X-ray crystallography is a widely used method, though in 50 years of work, it has only been possible to determine 18% of biological structures present in cells, due to difficulty crystallising biomolecules with large molecular weights. An alternative technique, cryo-electron microscopy (cryo-EM) overcomes this issue, allowing much larger structures to be described. In the next decade, it should be possible to image around 50% of subcellular structures. However, these larger length scales are beyond the capabilities of MD, as they would be many thousands of times too computationally expensive to compute such vast molecules atom-by-atom, even with High Performance Computing. Therefore, multi-scale multi-physics methods are required. For larger length scales, continuum mechanics is proposed for use in a coarse-grained approach, computing bulk quantities. Fluctuating Finite Element Analysis (FFEA) is a method primarily developed at the University of Leeds which uses meshes in place of atomic co-ordinates, therefore lending itself to data produced by cryo-EM [2]. With MD, scientists can currently study phenomena such as protein folding and molecular recognition. Modelling macromolecules at a larger scale, and exploring how they interact, will be the next step, helping to decipher the complex bio-machinery within cells. However, there are around 40 million proteins in a single cell, thus, even if only a small percentage interact, the computational demands still exceed the capabilities of serial programming, highlighting the need for massively parallel computing. This poster presents a mesh merging procedure that will be used to simulate protein-protein merging - one form of interaction, where two or more distinct proteins combine into one body. A methodology is proposed, whereby selected subsurfaces of two meshes are projected up to some defined plane, extruded, and then finally merged. The current developments are detailed, with techniques used to perform each stage explained, and a plan for future work is summarised. The described algorithm is now being incorporated into code_saturne [3], a multi-physics Computational Engineering code, due to the software’s massively parallel capabilities. The wider aim is to build a toolkit of algorithms to simulate the different protein interactions that occur in cells. It is hoped that these novel techniques will help to improve the predictive power of biosimulation, bridging the gap between molecular and clinical studies and helping to reduce costs of drug research. [1] Edwards et al., 2021. doi: 10.1107/S2059798321009712 [2] Solernou et al., 2018. doi: 10.1371/journal.pcbi.1005897 [3] Fournier et al., 2011. doi: 10.1016/j.compfluid.2011.01.028
Biomolecular simulations are crucial to understanding the complex and highly dynamic environments within biological cells. A popular approach relies on Molecular Dynamics (MD), which employs Newtonian mechanics and forcefields to compute atomic co-ordinates over short time steps. Biosimulations work in conjunction with experimental techniques, providing dynamic information to relate static biomolecular structures to functionality [1]. X-ray crystallography is a widely used method, though in 50 years of work, it has only been possible to determine 18% of biological structures present in cells, due to difficulty crystallising biomolecules with large molecular weights. An alternative technique, cryo-electron microscopy (cryo-EM) overcomes this issue, allowing much larger structures to be described. In the next decade, it should be possible to image around 50% of subcellular structures. However, these larger length scales are beyond the capabilities of MD, as they would be many thousands of times too computationally expensive to compute such vast molecules atom-by-atom, even with High Performance Computing. Therefore, multi-scale multi-physics methods are required. For larger length scales, continuum mechanics is proposed for use in a coarse-grained approach, computing bulk quantities. Fluctuating Finite Element Analysis (FFEA) is a method primarily developed at the University of Leeds which uses meshes in place of atomic co-ordinates, therefore lending itself to data produced by cryo-EM [2]. With MD, scientists can currently study phenomena such as protein folding and molecular recognition. Modelling macromolecules at a larger scale, and exploring how they interact, will be the next step, helping to decipher the complex bio-machinery within cells. However, there are around 40 million proteins in a single cell, thus, even if only a small percentage interact, the computational demands still exceed the capabilities of serial programming, highlighting the need for massively parallel computing. This poster presents a mesh merging procedure that will be used to simulate protein-protein merging - one form of interaction, where two or more distinct proteins combine into one body. A methodology is proposed, whereby selected subsurfaces of two meshes are projected up to some defined plane, extruded, and then finally merged. The current developments are detailed, with techniques used to perform each stage explained, and a plan for future work is summarised. The described algorithm is now being incorporated into code_saturne [3], a multi-physics Computational Engineering code, due to the software’s massively parallel capabilities. The wider aim is to build a toolkit of algorithms to simulate the different protein interactions that occur in cells. It is hoped that these novel techniques will help to improve the predictive power of biosimulation, bridging the gap between molecular and clinical studies and helping to reduce costs of drug research. [1] Edwards et al., 2021. doi: 10.1107/S2059798321009712 [2] Solernou et al., 2018. doi: 10.1371/journal.pcbi.1005897 [3] Fournier et al., 2011. doi: 10.1016/j.compfluid.2011.01.028
Format
On-site

